Fold Puzzle Database 2.0
Fuzzle 2.0 is a database of evolutionary related protein fragments.
Now with ligand information.

Protein evolution
Find evolutionary relationships between any protein of interest and the whole protein space. We have computed all-against-all sequence alignments for the proteins in the SCOPe database using profile hidden Markov state model comparisons.

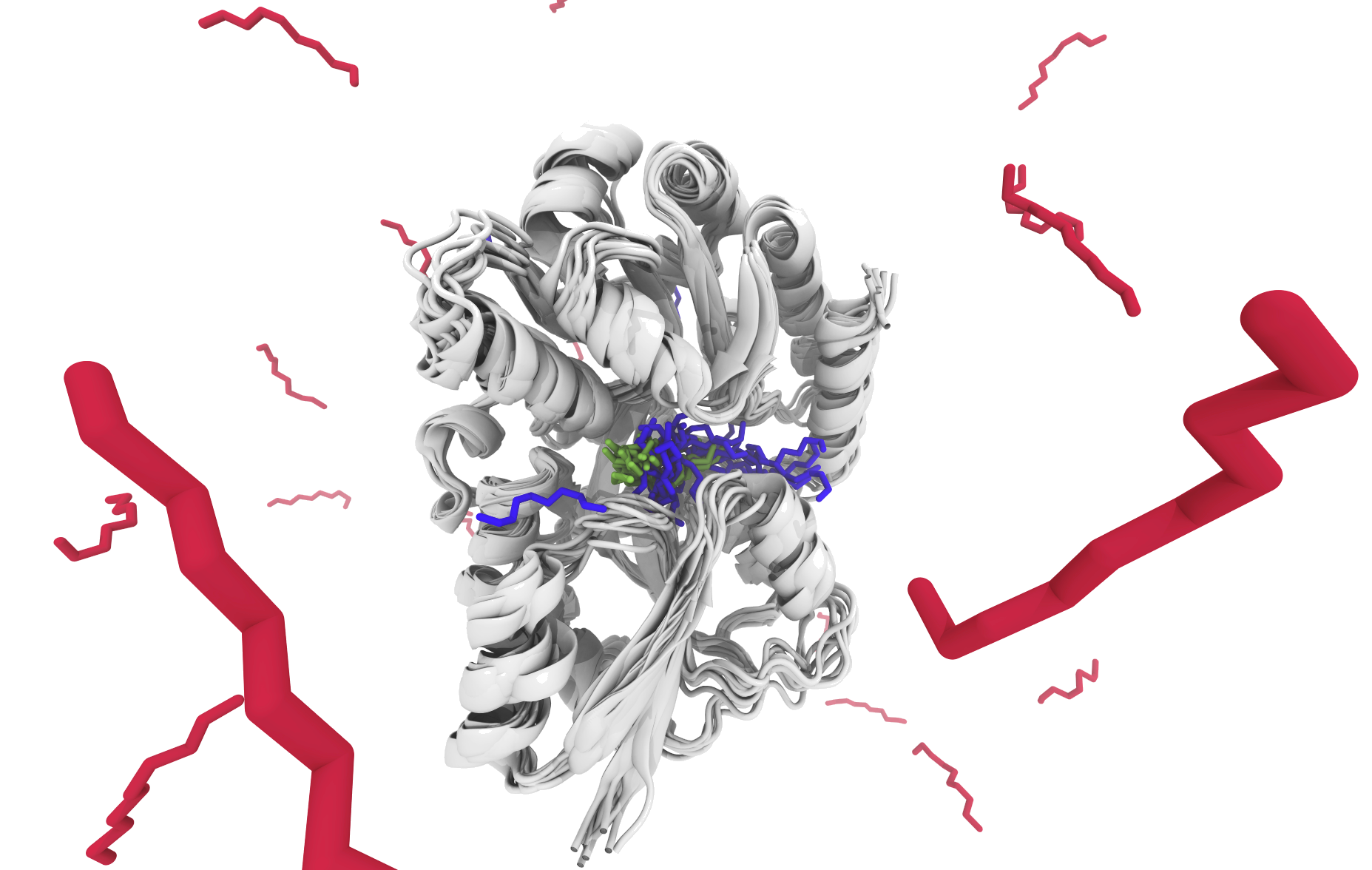
Ligand Binding
Find your ligand of interest in pockets of different proteins. You can visualize different interacting residues that the ligands bind to. You can also look for conserved protein fragments that bind your ligand and compare it to other ligands that also bind this fragment.

Relationships between folds
Conserved fragments can appear within the same family, superfamily or fold. In Fuzzle you can find conserved fragments across all SCOP categories.
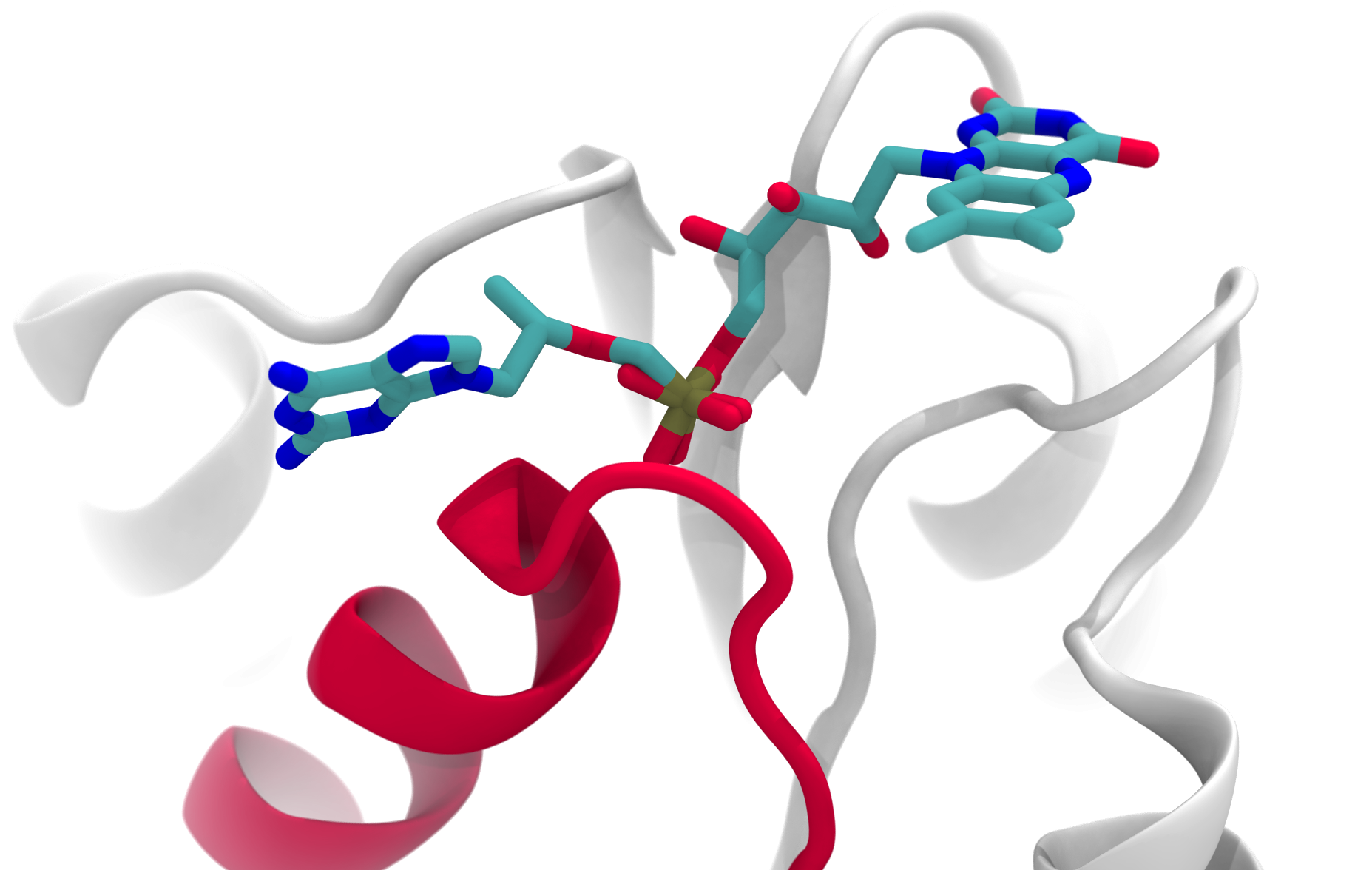
Conserved fragments. Do they contain conserved binding modes?
We found over 1000 protein fragments that appear across protein space. These fragments occur in proteins that belong to different folds (different structural topologies), yet they have retained a similar sequence and structure over the course of evolution. These fragments are often involved in ligand or cofactor binding, and sometimes it is precisely this function that has been selected. View details »

Protein universe. How are folds related?
Protein space can be visualized by similarity networks. We represent all proteins in SCOPe 95 as nodes of a network. Two domains are linked when they have a fragment in common. View details »
Documentation. Any questions?
Take a look at our documentation whenever you have a question. We tried our best to provide detailed explanations, to describe how the database and the protein networks were created, and how evolutionarily conserved fragments were identified. Feel free to drop us a line if you have further questions or suggestions. View details »