Ligand Documentation
Adding ligands into the SCOPe pdbs
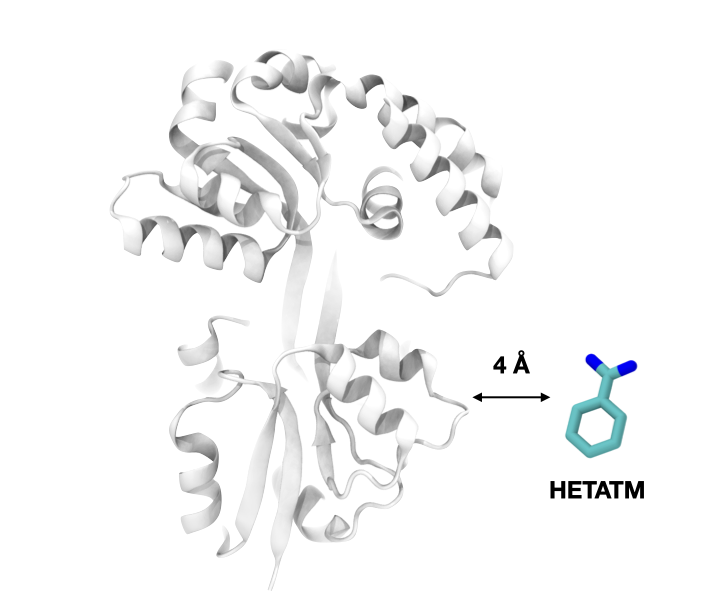
The SCOPe database classifies the structures from the PDB database according to their structure and topological connections hierarchically. This process occurs via a combination of automation and manual curation, and in doing so the heteroatoms from the PDB structures are discarded.We wanted to analyze our fragments also in the context of their functions. To that end, we needed to include information on how the fragments interact with biologically relevant molecules such as cofactors, ions, metals, or drugs. We mapped each of the domains in SCOPe to its corresponding PDB and retrieved those non-protein molecules that are within 4 Å of any domain atom. This way, we preserve the domain definition from SCOPe and include the ligand information from the original PDB.
Searching ligands in Fuzzle
PDB search
Ligands can be searched using their PDB identifier. Each ligand that appears in any PDB X-ray structure presents a unique three-letter code identifier (or two and one-letter identifier for ions and atoms), such as HEM (Heme group). The output will be a table with the representative domains that bind that ligand. It is also possible to retrieve all domains that bind this ligand, not only those that are representatives of their clusters in SCOPe95.SMILE search
Ligands can also be searched via their SMILE. SMILES are representations of ligand structures in the form of a line notation. They uniquely describe a ligand and can be reverted to their 2D structures, or can be used to quickly search similar compounds.- It is possible to find in Fuzzle all domains that exactly bind one ligand via its smile. Such as for example all representative domains that bind phosphate.
- It is also possible to search ligands that are at least 70% similar to a SMILE query, like all domains that bind compounds 70% similar to Flavin Mononucleotide.
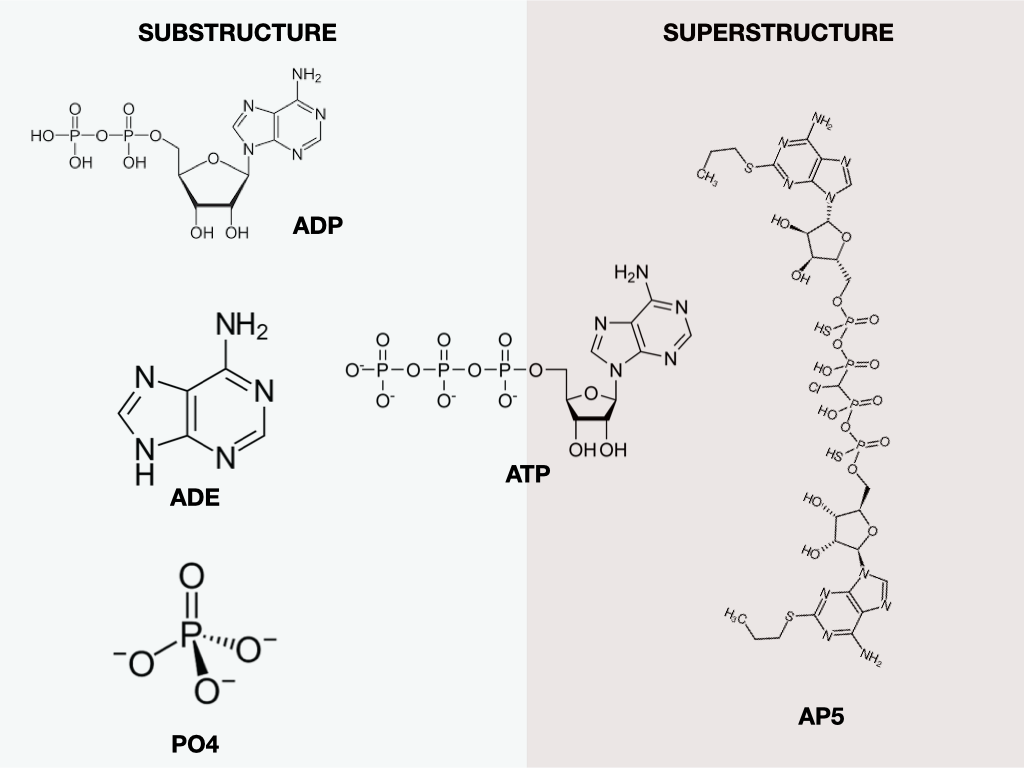
- Substructures and superstructures are also searchable. Superstructures are all those ligands that contain the query molecule as a moiety. For example, ATP is a superstructure of a phosphate molecule or ADP. On the other hand, a substructure search will find all moieties contained within the query molecule. An example is a benzene molecule, which is a substructure of the molecule indole (and many other compounds). The figure below exemplifies the ATP example.