Background
Previous work
Proteins are critical biological components that take part in all cellular processes. When we look at protein structures we can identify parts that form separated folding units, termed domains. To put an example, we find the same domain in white in this figure:
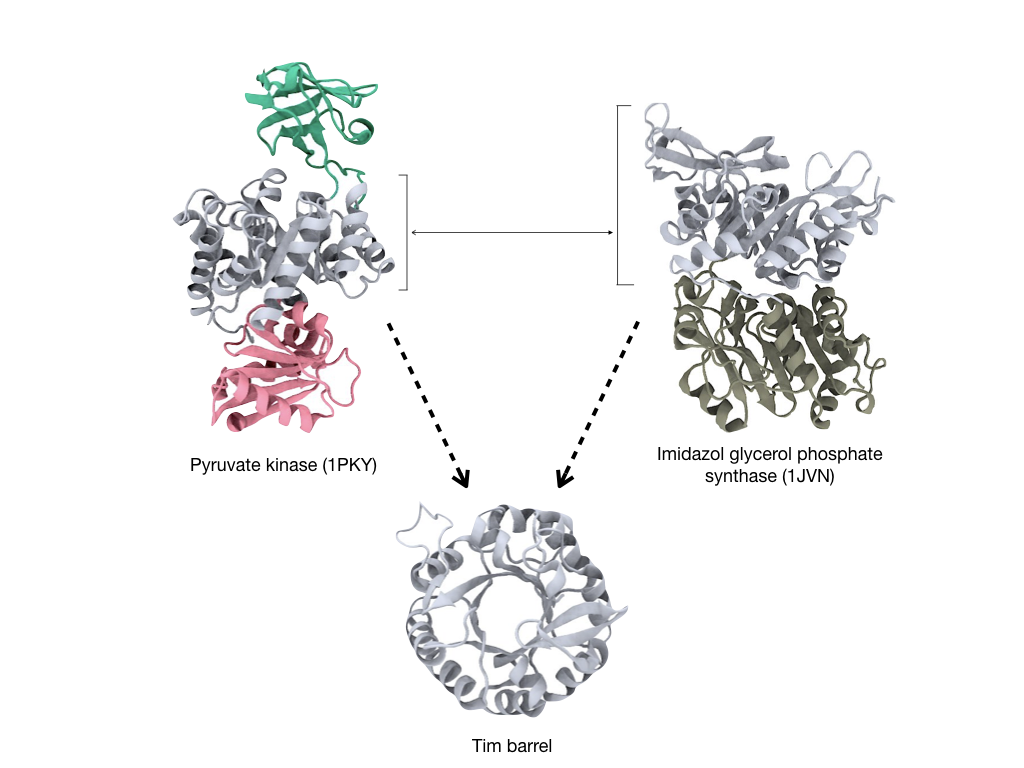
The TIM-barrel or (βα)8-barrel appears both in the pyruvate kinase and Imidazol glycerol phosphate synthase proteins. It is commonly understood that proteins evolved through the recruitment of domains and for this reason domains have been for long been defined as the basic evolutionary unit.
When we look at the domains themselves, we can distinguish smaller fragments from which the domains are formed. For example, in this figure we can see that the TIM-barrel and the flavodoxin domains, two ancient and highly populated superfolds, share a fragment:
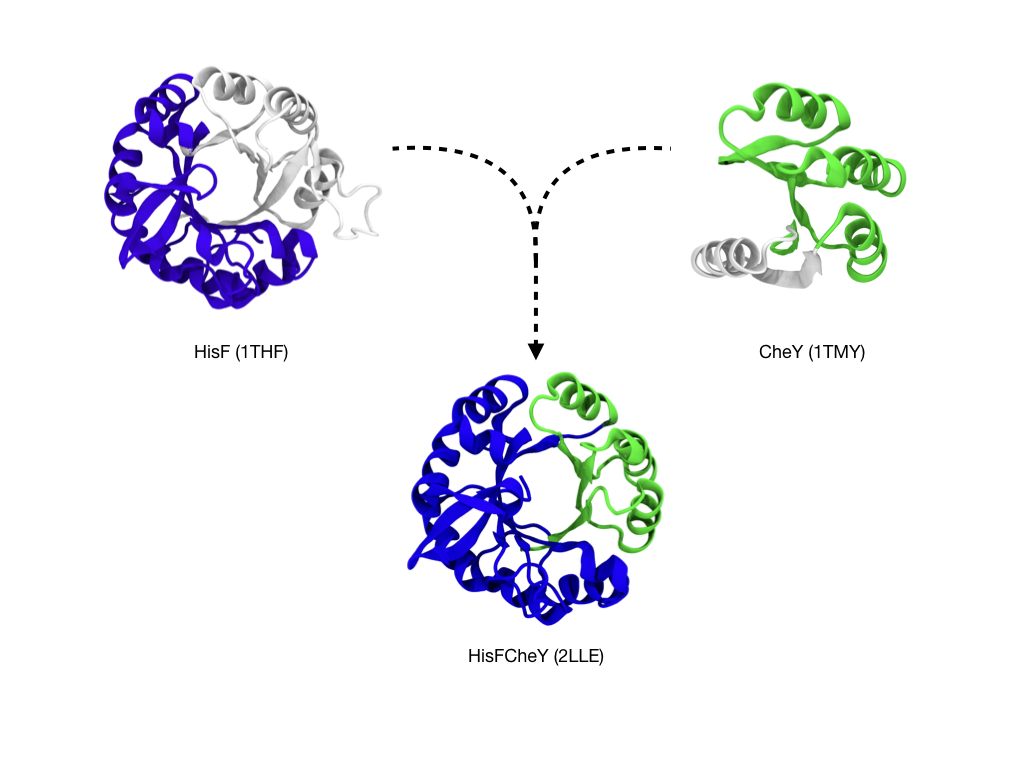
By using this structural evidence, a chimeric protein made from CheY and HisF was built
from smaller fragments.
The resulting protein formed a barrel-like structure [1, 2], like in the above figure.
In a follow-up study it was investigated whether these two proteins that had been chosen based
on structural evidence were in fact evolutionary related. To do so, a search using profile
hidden markov model (HMM) comparisons, able to find remote sequence homology,
revealed strong evidence of homology between
proteins of these two ancient and highly populated folds. Furthermore, a family of sequences
that show intermediate features between both folds was detected, enabling the first crystallization
of one of its members [3]:
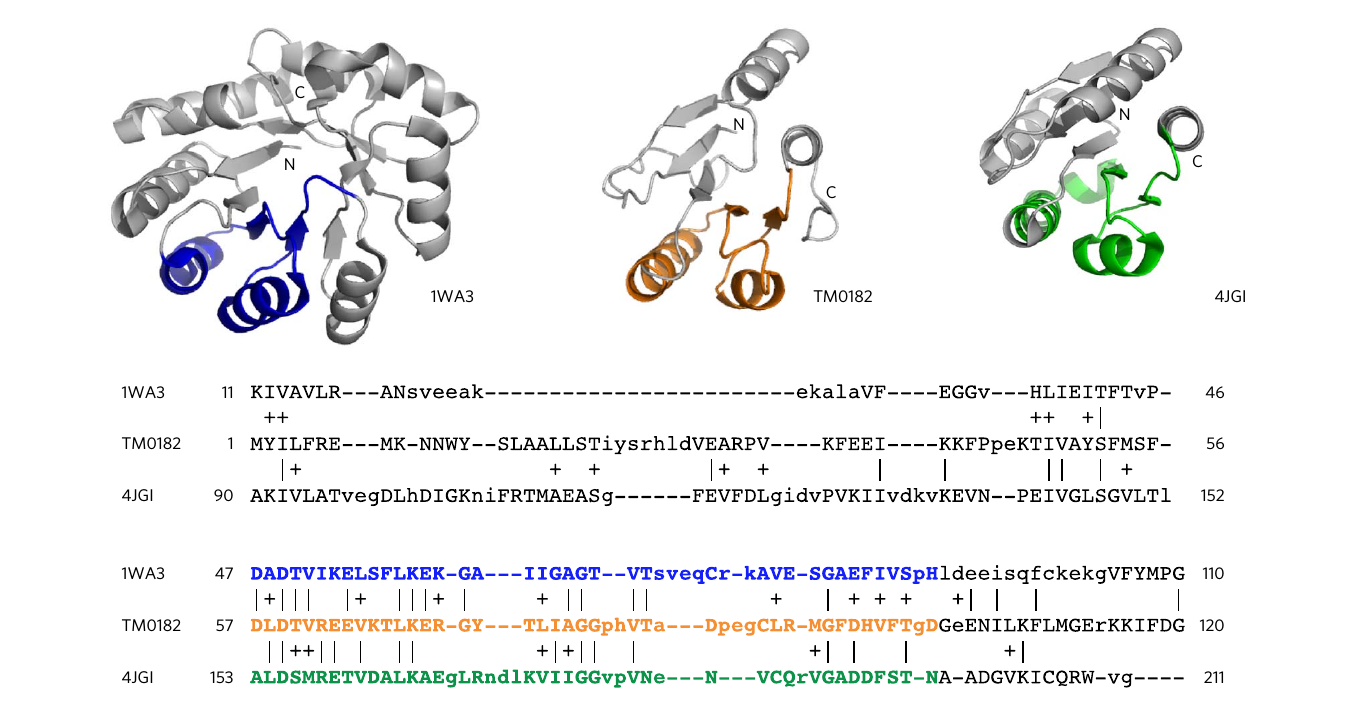
These results support the vision that protein folds share a common ancestry and that sequence similarity searches such as the one in this work are useful tools to identify sub-domain sized fragments that, mimicking Nature, can serve as building blocks. We extended the previous search between the TIM-barrel and flavodoxin folds to a global search of the protein sequence space and stored here in FUZZLE. You can get detailed information on how the database was built here.
References
[1] TAM Bharat, S Eisenbeis, K Zeth, B Höcker A βα-barrel built by the combination of fragments from different folds. Proceedings of the National Academy of Sciences
[2] Simone Eisenbeis, William Proffitt, Murray Coles, Vincent Truffault, Sooruban Shanmugaratnam, Jens Meiler, Birte Höcker. Potential of fragment recombination for rational design of proteins. Journal of the American Chemical Society 134 (9), 4019-4022
[3] JA Farías-Rico, S Schmidt, B Höcker. Evolutionary relationship of two ancient protein superfolds. Nature chemical biology 10 (9), 710